Cervical Cancer Risk Classification
library(tidyverse)
library(Boruta)
library(skimr)
library(DT)
library(skimr)
library(janitor)
library(hrbrthemes)
library(tidylog)
library(vip)
library(broom)
library(GGally)
library(cowplot)
theme_set(theme_ipsum_ps())
Cervical cancer risk clasification dataset:
https://www.kaggle.com/loveall/img/posts/cervical-cancer-risk-classification
Original dataset
raw.data <-
read_csv("kag_risk_factors_cervical_cancer.csv")
# raw.data %>%
# datatable()
Changing column names to BigCamelCase, converting ‘?’ to ‘NA’ values, removing empty rows and columns
raw.data <-
raw.data %>%
clean_names(case='big_camel') %>%
na_if('?') %>%
remove_empty(c("rows", "cols"))
glimpse(raw.data)
## Rows: 858
## Columns: 36
## $ Age <dbl> 18, 15, 34, 52, 46, 42, 51, 26, 45, 4…
## $ NumberOfSexualPartners <chr> "4.0", "1.0", "1.0", "5.0", "3.0", "3…
## $ FirstSexualIntercourse <chr> "15.0", "14.0", NA, "16.0", "21.0", "…
## $ NumOfPregnancies <chr> "1.0", "1.0", "1.0", "4.0", "4.0", "2…
## $ Smokes <chr> "0.0", "0.0", "0.0", "1.0", "0.0", "0…
## $ SmokesYears <chr> "0.0", "0.0", "0.0", "37.0", "0.0", "…
## $ SmokesPacksYear <chr> "0.0", "0.0", "0.0", "37.0", "0.0", "…
## $ HormonalContraceptives <chr> "0.0", "0.0", "0.0", "1.0", "1.0", "0…
## $ HormonalContraceptivesYears <chr> "0.0", "0.0", "0.0", "3.0", "15.0", "…
## $ Iud <chr> "0.0", "0.0", "0.0", "0.0", "0.0", "0…
## $ IudYears <chr> "0.0", "0.0", "0.0", "0.0", "0.0", "0…
## $ StDs <chr> "0.0", "0.0", "0.0", "0.0", "0.0", "0…
## $ StDsNumber <chr> "0.0", "0.0", "0.0", "0.0", "0.0", "0…
## $ StDsCondylomatosis <chr> "0.0", "0.0", "0.0", "0.0", "0.0", "0…
## $ StDsCervicalCondylomatosis <chr> "0.0", "0.0", "0.0", "0.0", "0.0", "0…
## $ StDsVaginalCondylomatosis <chr> "0.0", "0.0", "0.0", "0.0", "0.0", "0…
## $ StDsVulvoPerinealCondylomatosis <chr> "0.0", "0.0", "0.0", "0.0", "0.0", "0…
## $ StDsSyphilis <chr> "0.0", "0.0", "0.0", "0.0", "0.0", "0…
## $ StDsPelvicInflammatoryDisease <chr> "0.0", "0.0", "0.0", "0.0", "0.0", "0…
## $ StDsGenitalHerpes <chr> "0.0", "0.0", "0.0", "0.0", "0.0", "0…
## $ StDsMolluscumContagiosum <chr> "0.0", "0.0", "0.0", "0.0", "0.0", "0…
## $ StDsAids <chr> "0.0", "0.0", "0.0", "0.0", "0.0", "0…
## $ StDsHiv <chr> "0.0", "0.0", "0.0", "0.0", "0.0", "0…
## $ StDsHepatitisB <chr> "0.0", "0.0", "0.0", "0.0", "0.0", "0…
## $ StDsHpv <chr> "0.0", "0.0", "0.0", "0.0", "0.0", "0…
## $ StDsNumberOfDiagnosis <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
## $ StDsTimeSinceFirstDiagnosis <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
## $ StDsTimeSinceLastDiagnosis <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
## $ DxCancer <dbl> 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0…
## $ DxCin <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
## $ DxHpv <dbl> 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0…
## $ Dx <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0…
## $ Hinselmann <dbl> 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0…
## $ Schiller <dbl> 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0…
## $ Citology <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
## $ Biopsy <dbl> 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0…
Converting datatype to numeric or factor whichever suitable
facotrs for those columns that can be grouped under common subject
raw.data <-
raw.data %>%
mutate_all(as.numeric) %>%
mutate(Cancer=(Hinselmann+Schiller+Citology+Biopsy)>0) %>%
select(-(Hinselmann:Biopsy)) %>%
mutate(SmokeAmount=SmokesYears*SmokesPacksYear) %>%
select(-SmokesYears,-SmokesPacksYear) %>%
mutate_if(is.numeric, ~round(., 1)) %>%
mutate(across(c(NumberOfSexualPartners,
NumOfPregnancies,Smokes,
HormonalContraceptives,
Iud,
StDs,
StDsNumber,
StDsCondylomatosis,
StDsCervicalCondylomatosis,
StDsVaginalCondylomatosis,
StDsVulvoPerinealCondylomatosis,
StDsSyphilis,
StDsPelvicInflammatoryDisease,
StDsGenitalHerpes,
StDsMolluscumContagiosum,
StDsAids,
StDsHiv,
StDsHepatitisB,
StDsHpv,
StDsNumberOfDiagnosis,
DxCancer,
DxCin,
DxHpv,
Dx,
Cancer),as.factor))
glimpse(raw.data)
## Rows: 858
## Columns: 32
## $ Age <dbl> 18, 15, 34, 52, 46, 42, 51, 26, 45, 4…
## $ NumberOfSexualPartners <fct> 4, 1, 1, 5, 3, 3, 3, 1, 1, 3, 3, 1, 4…
## $ FirstSexualIntercourse <dbl> 15, 14, NA, 16, 21, 23, 17, 26, 20, 1…
## $ NumOfPregnancies <fct> 1, 1, 1, 4, 4, 2, 6, 3, 5, NA, 4, 3, …
## $ Smokes <fct> 0, 0, 0, 1, 0, 0, 1, 0, 0, 1, 0, 0, 0…
## $ HormonalContraceptives <fct> 0, 0, 0, 1, 1, 0, 0, 1, 0, 0, 1, 1, 1…
## $ HormonalContraceptivesYears <dbl> 0.0, 0.0, 0.0, 3.0, 15.0, 0.0, 0.0, 2…
## $ Iud <fct> 0, 0, 0, 0, 0, 0, 1, 1, 0, NA, 0, 0, …
## $ IudYears <dbl> 0, 0, 0, 0, 0, 0, 7, 7, 0, NA, 0, 0, …
## $ StDs <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
## $ StDsNumber <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
## $ StDsCondylomatosis <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
## $ StDsCervicalCondylomatosis <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
## $ StDsVaginalCondylomatosis <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
## $ StDsVulvoPerinealCondylomatosis <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
## $ StDsSyphilis <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
## $ StDsPelvicInflammatoryDisease <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
## $ StDsGenitalHerpes <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
## $ StDsMolluscumContagiosum <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
## $ StDsAids <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
## $ StDsHiv <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
## $ StDsHepatitisB <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
## $ StDsHpv <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
## $ StDsNumberOfDiagnosis <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
## $ StDsTimeSinceFirstDiagnosis <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
## $ StDsTimeSinceLastDiagnosis <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
## $ DxCancer <fct> 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0…
## $ DxCin <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
## $ DxHpv <fct> 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0…
## $ Dx <fct> 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0…
## $ Cancer <fct> FALSE, FALSE, FALSE, FALSE, FALSE, FA…
## $ SmokeAmount <dbl> 0.0, 0.0, 0.0, 1369.0, 0.0, 0.0, 115.…
raw.data %>%
skim()
| Name | Piped data |
| Number of rows | 858 |
| Number of columns | 32 |
| _______________________ | |
| Column type frequency: | |
| factor | 25 |
| numeric | 7 |
| ________________________ | |
| Group variables | None |
Data summary
Variable type: factor
| skim_variable | n_missing | complete_rate | ordered | n_unique | top_counts |
|---|---|---|---|---|---|
| NumberOfSexualPartners | 26 | 0.97 | FALSE | 12 | 2: 272, 3: 208, 1: 206, 4: 78 |
| NumOfPregnancies | 56 | 0.93 | FALSE | 11 | 1: 270, 2: 240, 3: 139, 4: 74 |
| Smokes | 13 | 0.98 | FALSE | 2 | 0: 722, 1: 123 |
| HormonalContraceptives | 108 | 0.87 | FALSE | 2 | 1: 481, 0: 269 |
| Iud | 117 | 0.86 | FALSE | 2 | 0: 658, 1: 83 |
| StDs | 105 | 0.88 | FALSE | 2 | 0: 674, 1: 79 |
| StDsNumber | 105 | 0.88 | FALSE | 5 | 0: 674, 2: 37, 1: 34, 3: 7 |
| StDsCondylomatosis | 105 | 0.88 | FALSE | 2 | 0: 709, 1: 44 |
| StDsCervicalCondylomatosis | 105 | 0.88 | FALSE | 1 | 0: 753 |
| StDsVaginalCondylomatosis | 105 | 0.88 | FALSE | 2 | 0: 749, 1: 4 |
| StDsVulvoPerinealCondylomatosis | 105 | 0.88 | FALSE | 2 | 0: 710, 1: 43 |
| StDsSyphilis | 105 | 0.88 | FALSE | 2 | 0: 735, 1: 18 |
| StDsPelvicInflammatoryDisease | 105 | 0.88 | FALSE | 2 | 0: 752, 1: 1 |
| StDsGenitalHerpes | 105 | 0.88 | FALSE | 2 | 0: 752, 1: 1 |
| StDsMolluscumContagiosum | 105 | 0.88 | FALSE | 2 | 0: 752, 1: 1 |
| StDsAids | 105 | 0.88 | FALSE | 1 | 0: 753 |
| StDsHiv | 105 | 0.88 | FALSE | 2 | 0: 735, 1: 18 |
| StDsHepatitisB | 105 | 0.88 | FALSE | 2 | 0: 752, 1: 1 |
| StDsHpv | 105 | 0.88 | FALSE | 2 | 0: 751, 1: 2 |
| StDsNumberOfDiagnosis | 0 | 1.00 | FALSE | 4 | 0: 787, 1: 68, 2: 2, 3: 1 |
| DxCancer | 0 | 1.00 | FALSE | 2 | 0: 840, 1: 18 |
| DxCin | 0 | 1.00 | FALSE | 2 | 0: 849, 1: 9 |
| DxHpv | 0 | 1.00 | FALSE | 2 | 0: 840, 1: 18 |
| Dx | 0 | 1.00 | FALSE | 2 | 0: 834, 1: 24 |
| Cancer | 0 | 1.00 | FALSE | 2 | FAL: 756, TRU: 102 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| Age | 0 | 1.00 | 26.82 | 8.50 | 13 | 20 | 25.0 | 32.0 | 84 | ▇▅▁▁▁ |
| FirstSexualIntercourse | 7 | 0.99 | 17.00 | 2.80 | 10 | 15 | 17.0 | 18.0 | 32 | ▂▇▂▁▁ |
| HormonalContraceptivesYears | 108 | 0.87 | 2.26 | 3.76 | 0 | 0 | 0.5 | 3.0 | 30 | ▇▁▁▁▁ |
| IudYears | 117 | 0.86 | 0.51 | 1.94 | 0 | 0 | 0.0 | 0.0 | 19 | ▇▁▁▁▁ |
| StDsTimeSinceFirstDiagnosis | 787 | 0.08 | 6.14 | 5.90 | 1 | 2 | 4.0 | 8.0 | 22 | ▇▂▁▁▁ |
| StDsTimeSinceLastDiagnosis | 787 | 0.08 | 5.82 | 5.76 | 1 | 2 | 3.0 | 7.5 | 22 | ▇▂▁▁▁ |
| SmokeAmount | 13 | 0.98 | 7.14 | 57.29 | 0 | 0 | 0.0 | 0.0 | 1369 | ▇▁▁▁▁ |
Plots
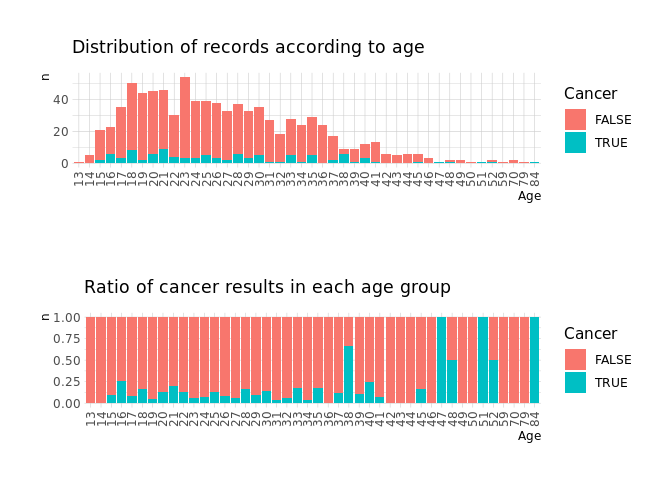
Age
age1 <-
raw.data %>%
count(Age,Cancer) %>%
mutate(Age=factor(Age)) %>%
ggplot(aes(Age,n,fill=Cancer))+
geom_col()+
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))+
labs(subtitle = 'Distribution of records according to age')
age2 <-
raw.data %>%
count(Age,Cancer) %>%
mutate(Age=factor(Age)) %>%
ggplot(aes(Age,n,fill=Cancer))+
geom_col(position='fill')+
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))+
labs(subtitle='Ratio of cancer results in each age group')
cowplot::plot_grid(age1, age2, nrow=2)
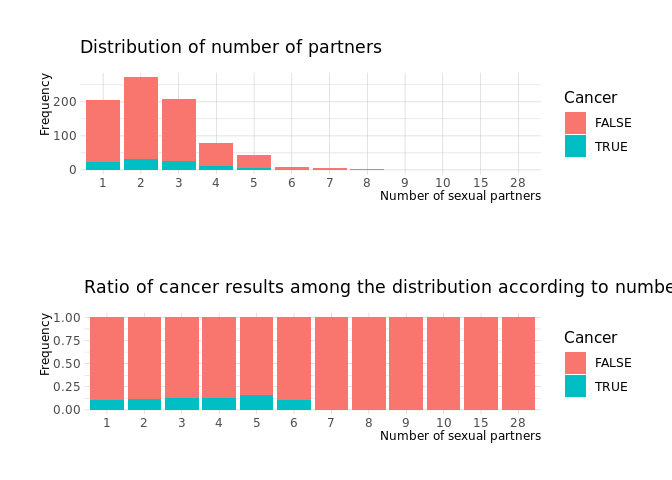
Number of sexual partners
no_of_sexual_partners_1 <-
raw.data %>%
count(NumberOfSexualPartners,Cancer) %>%
filter(!is.na(NumberOfSexualPartners)) %>%
group_by(NumberOfSexualPartners) %>%
ggplot(aes(x=NumberOfSexualPartners, y=n,fill=Cancer)) +
geom_col()+
labs(
x = 'Number of sexual partners',
y = 'Frequency',
subtitle= 'Distribution of number of partners'
)
no_of_sexual_partners_2 <-
raw.data %>%
count(NumberOfSexualPartners,Cancer) %>%
filter(!is.na(NumberOfSexualPartners)) %>%
group_by(NumberOfSexualPartners) %>%
ggplot(aes(x=NumberOfSexualPartners, y=n,fill=Cancer)) +
geom_col(position = 'fill')+
labs(
x = 'Number of sexual partners',
y = 'Frequency',
subtitle= 'Ratio of cancer results among the distribution according to number of partners'
)
cowplot::plot_grid(no_of_sexual_partners_1,no_of_sexual_partners_2, nrow=2)
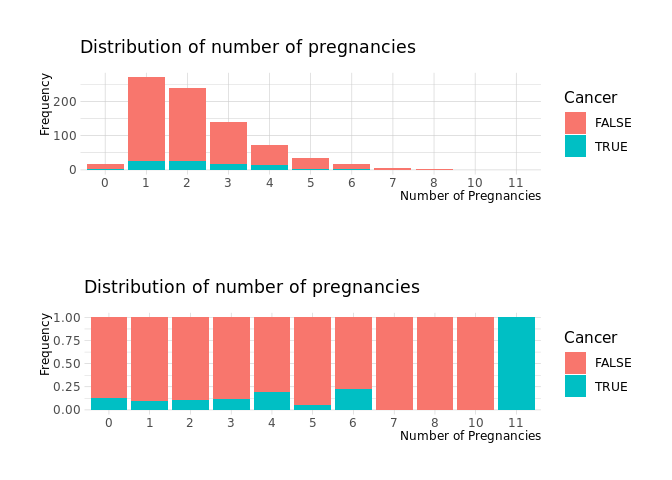
Number of pregnancies
no_of_pregnancies_1 <-
raw.data %>%
count(NumOfPregnancies,Cancer) %>%
filter(!is.na(NumOfPregnancies)) %>%
ggplot(aes(x=NumOfPregnancies, y=n,fill=Cancer)) +
geom_col()+
labs(
x = 'Number of Pregnancies',
y = 'Frequency',
subtitle= 'Distribution of number of pregnancies'
)
no_of_pregnancies_2 <-
raw.data %>%
count(NumOfPregnancies,Cancer) %>%
filter(!is.na(NumOfPregnancies)) %>%
ggplot(aes(x=NumOfPregnancies, y=n,fill=Cancer)) +
geom_col(position = 'fill')+
labs(
x = 'Number of Pregnancies',
y = 'Frequency',
subtitle= 'Distribution of number of pregnancies'
)
cowplot::plot_grid(no_of_pregnancies_1,no_of_pregnancies_2, nrow=2)
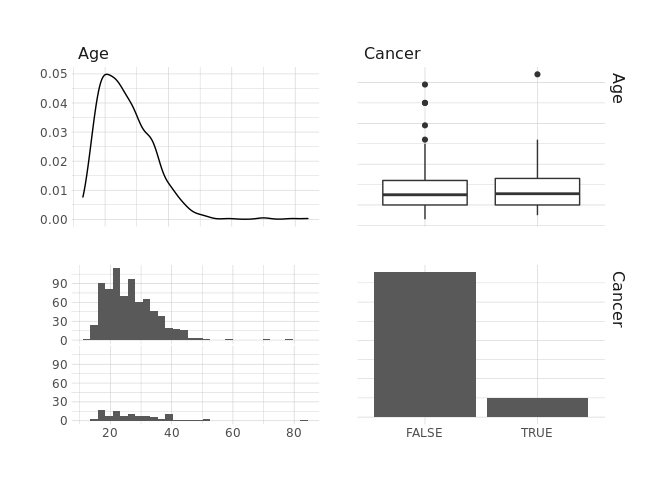
GGPairs
raw.data %>%
select(Age,Cancer) %>%
ggpairs()
raw.data %>%
select(NumberOfSexualPartners,Cancer) %>%
ggpairs()
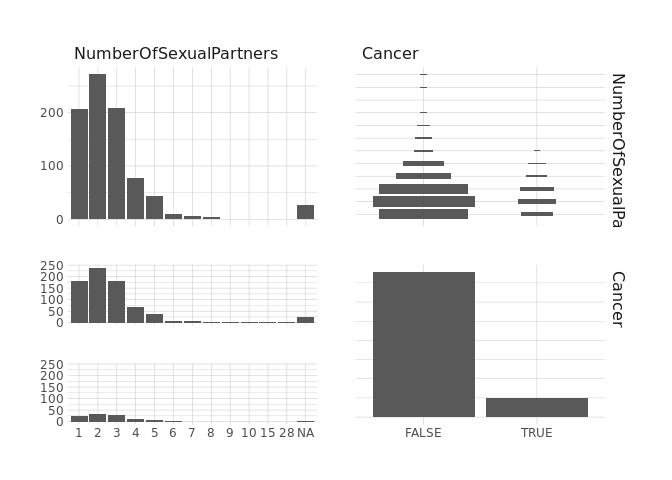
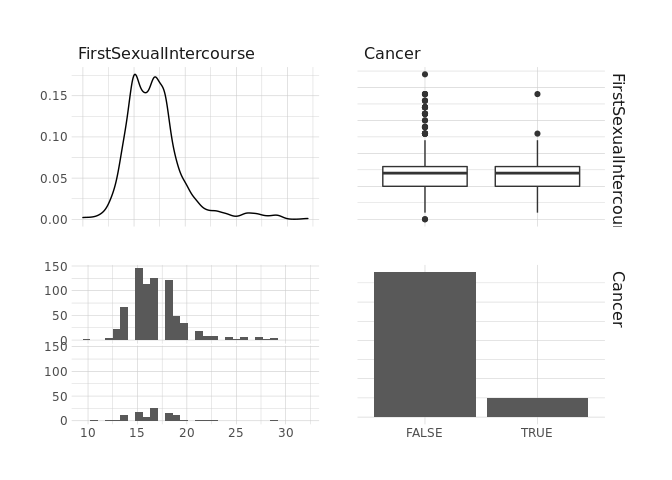
raw.data %>%
select(FirstSexualIntercourse,Cancer) %>%
ggpairs()
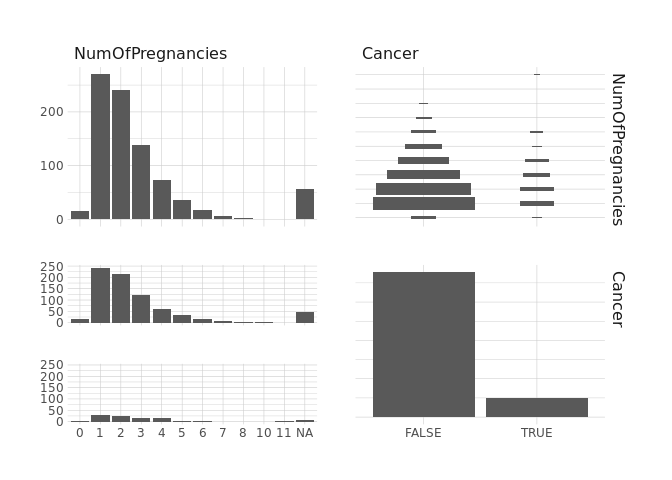
raw.data %>%
select(NumOfPregnancies,Cancer) %>%
ggpairs()
raw.data %>%
select(Smokes,Cancer) %>%
ggpairs()
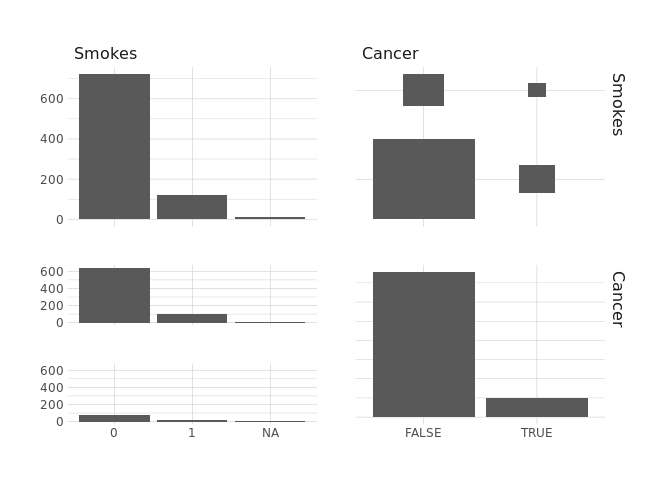
raw.data %>%
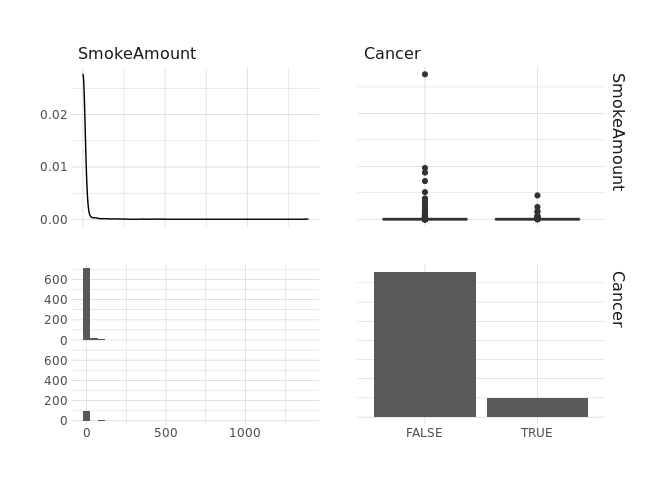
select(SmokeAmount,Cancer) %>%
ggpairs()
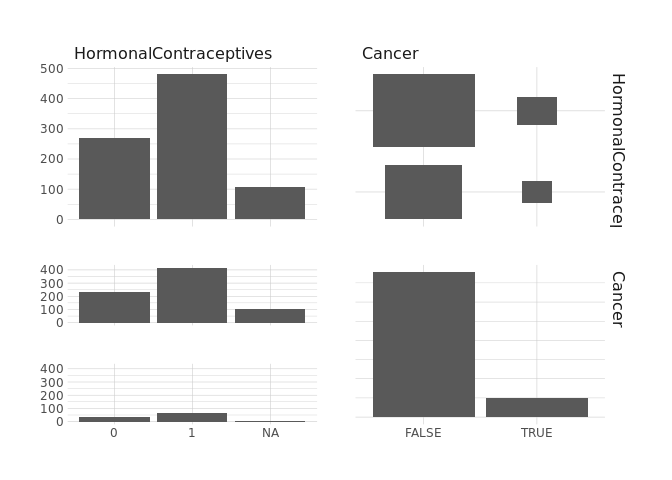
raw.data %>%
select(HormonalContraceptives,Cancer) %>%
ggpairs()
raw.data %>%
select(Iud,Cancer) %>%
ggpairs()
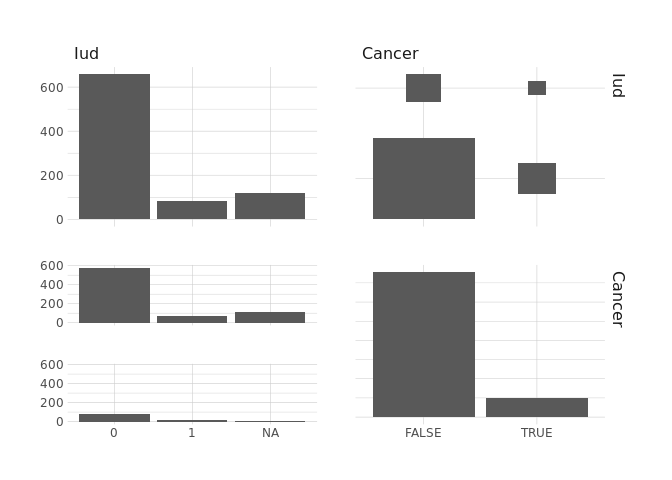
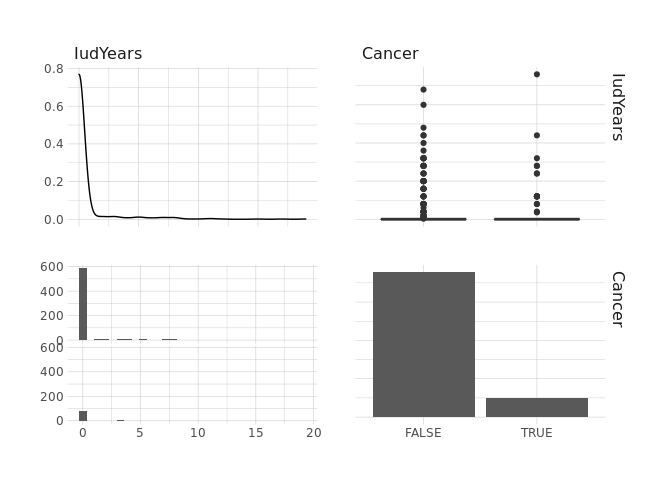
raw.data %>%
select(IudYears,Cancer) %>%
ggpairs()
raw.data %>%
select(StDs,Cancer) %>%
ggpairs()
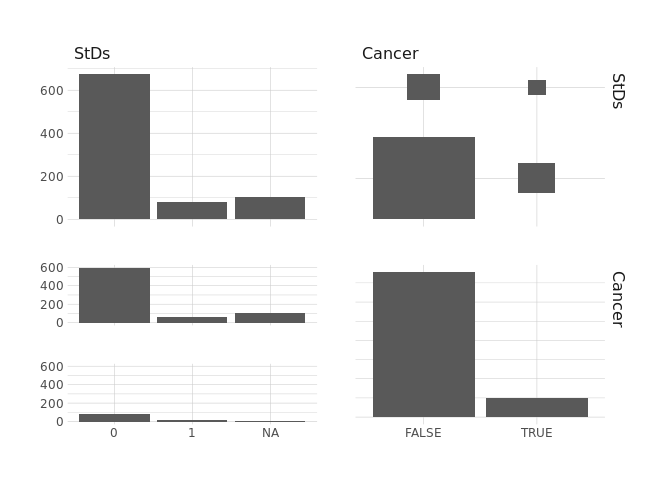
raw.data %>%
select(StDsNumber,Cancer) %>%
ggpairs()
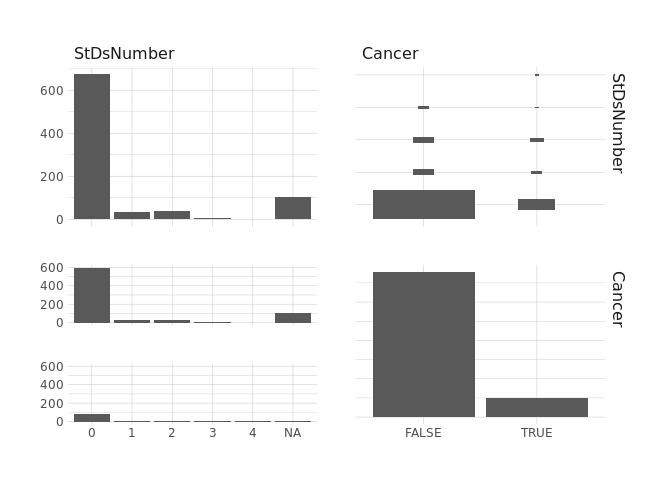
raw.data %>%
select(StDsCondylomatosis,Cancer) %>%
ggpairs()
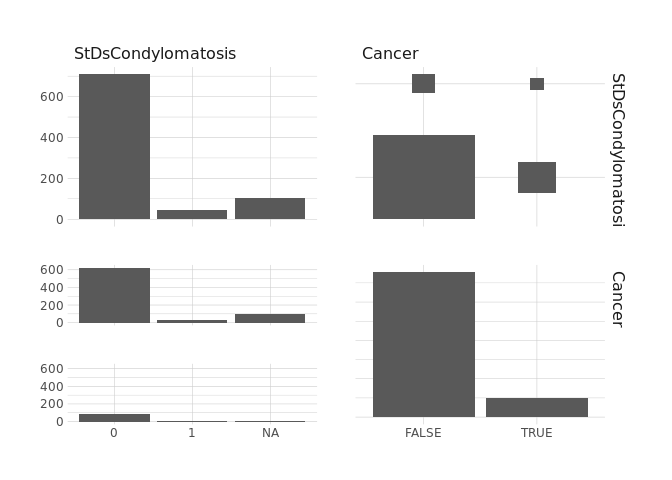
raw.data %>%
select(StDsCervicalCondylomatosis,Cancer) %>%
ggpairs()
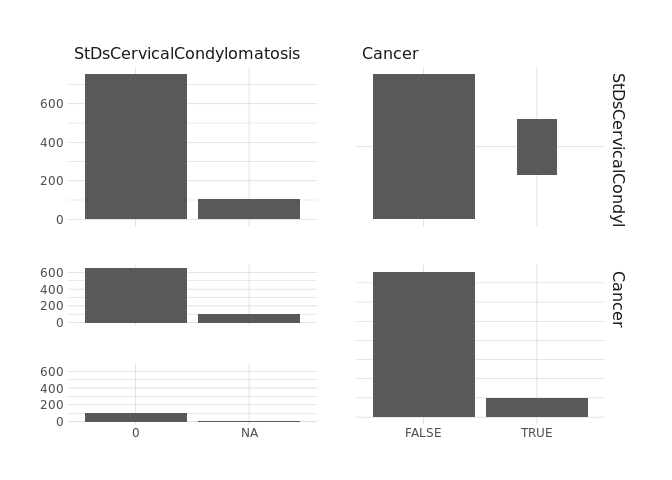
raw.data %>%
select(StDsVaginalCondylomatosis,Cancer) %>%
ggpairs()
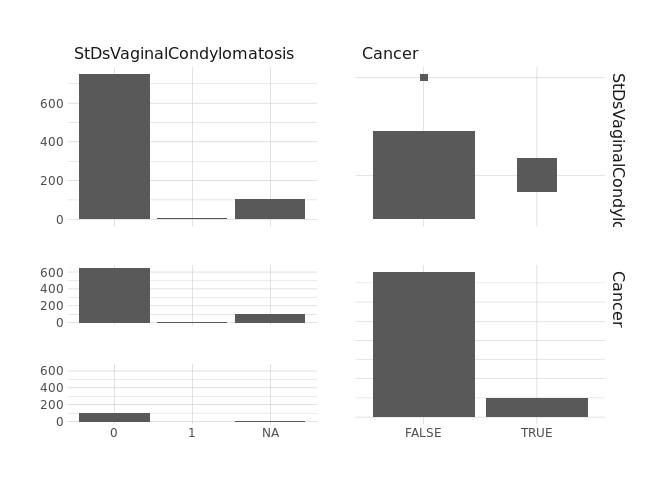
raw.data %>%
select(StDsVulvoPerinealCondylomatosis,Cancer) %>%
ggpairs()
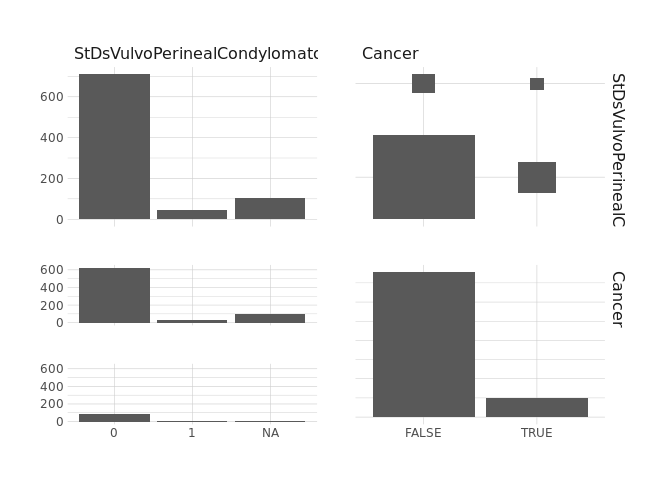
raw.data %>%
select(StDsNumberOfDiagnosis,Cancer) %>%
ggpairs()
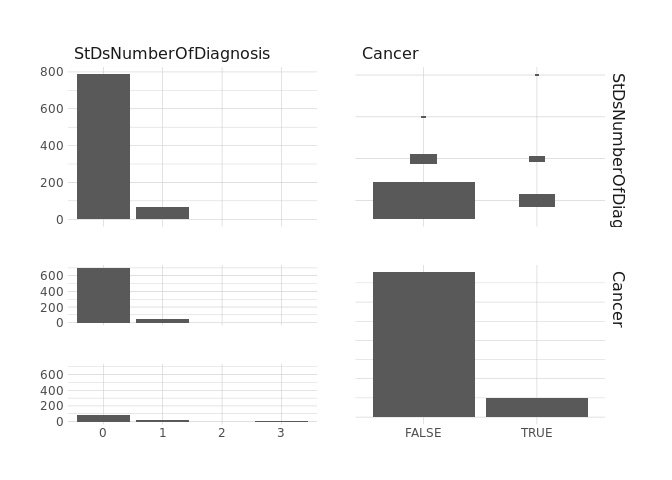
raw.data %>%
select(DxCancer,Cancer) %>%
ggpairs()
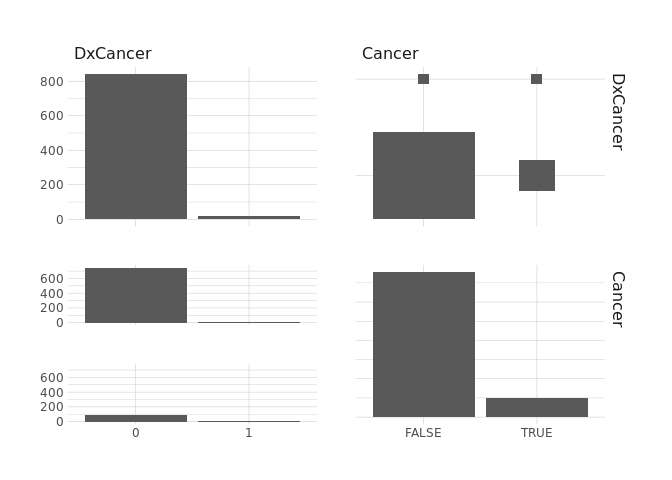
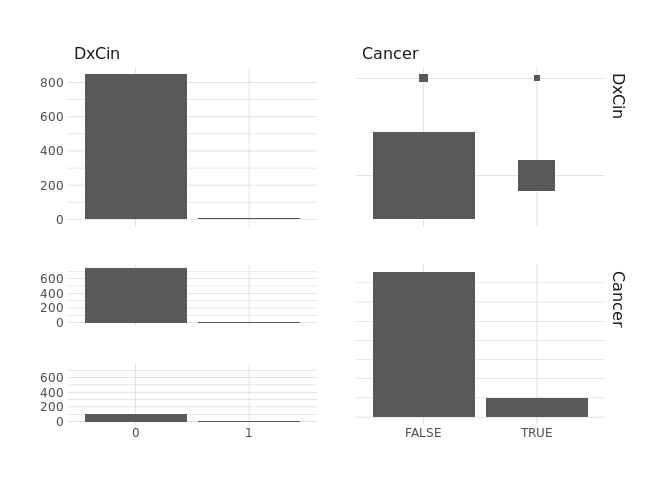
raw.data %>%
select(DxCin,Cancer) %>%
ggpairs()
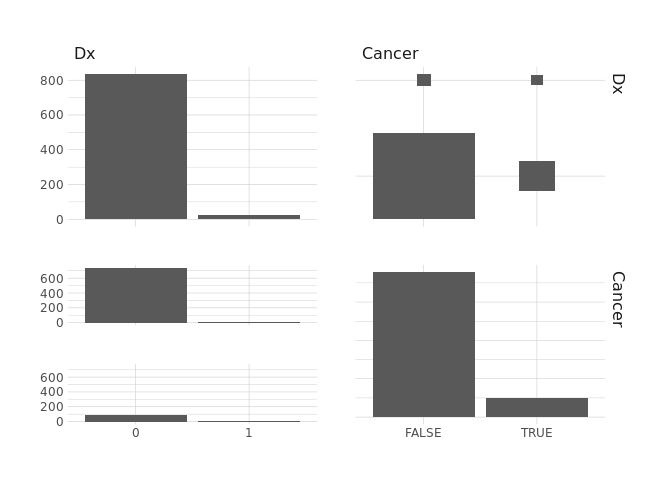
raw.data %>%
select(Dx,Cancer) %>%
ggpairs()
logistic regression model
new.data <-
raw.data %>%
select(Age,Smokes,Cancer)
model <-
glm(Cancer~Age+Smokes+Age*Smokes,new.data,family='binomial')
model %>%
tidy()
## # A tibble: 4 x 5
## term estimate std.error statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) -2.12 0.404 -5.24 0.000000160
## 2 Age 0.000702 0.0145 0.0486 0.961
## 3 Smokes1 -0.279 0.787 -0.355 0.723
## 4 Age:Smokes1 0.0275 0.0257 1.07 0.285
model %>%
glance()
## # A tibble: 1 x 8
## null.deviance df.null logLik AIC BIC deviance df.residual nobs
## <dbl> <int> <dbl> <dbl> <dbl> <dbl> <int> <int>
## 1 615. 844 -305. 617. 636. 609. 841 845
model %>%
vip()
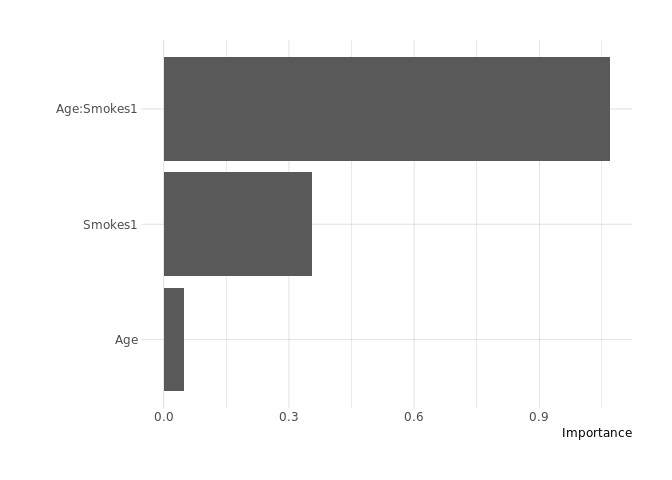
model %>%
vi()
## # A tibble: 3 x 3
## Variable Importance Sign
## <chr> <dbl> <chr>
## 1 Age:Smokes1 1.07 POS
## 2 Smokes1 0.355 NEG
## 3 Age 0.0486 POS
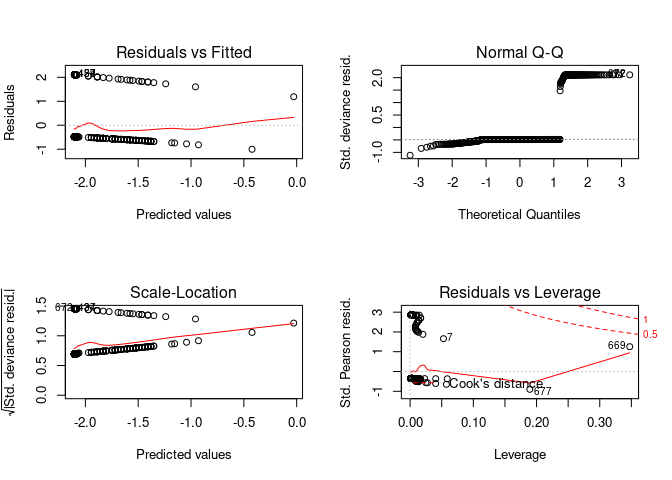
par(mfrow=c(2,2))
plot(model)
anova(model)
## Analysis of Deviance Table
##
## Model: binomial, link: logit
##
## Response: Cancer
##
## Terms added sequentially (first to last)
##
##
## Df Deviance Resid. Df Resid. Dev
## NULL 844 614.50
## Age 1 0.7209 843 613.78
## Smokes 1 3.2835 842 610.50
## Age:Smokes 1 1.1292 841 609.37
Prediction
test.data <-
new.data %>%
select(-Cancer)
final <-
tibble(predicted = predict(model,test.data), actual = new.data$Cancer) %>%
mutate(predicted = predicted>0)
final
## # A tibble: 858 x 2
## predicted actual
## <lgl> <fct>
## 1 FALSE FALSE
## 2 FALSE FALSE
## 3 FALSE FALSE
## 4 FALSE FALSE
## 5 FALSE FALSE
## 6 FALSE FALSE
## 7 FALSE TRUE
## 8 FALSE FALSE
## 9 FALSE FALSE
## 10 FALSE FALSE
## # … with 848 more rows
final %>%
filter(predicted == actual)
## # A tibble: 745 x 2
## predicted actual
## <lgl> <fct>
## 1 FALSE FALSE
## 2 FALSE FALSE
## 3 FALSE FALSE
## 4 FALSE FALSE
## 5 FALSE FALSE
## 6 FALSE FALSE
## 7 FALSE FALSE
## 8 FALSE FALSE
## 9 FALSE FALSE
## 10 FALSE FALSE
## # … with 735 more rows
final %>%
filter(predicted != actual)
## # A tibble: 100 x 2
## predicted actual
## <lgl> <fct>
## 1 FALSE TRUE
## 2 FALSE TRUE
## 3 FALSE TRUE
## 4 FALSE TRUE
## 5 FALSE TRUE
## 6 FALSE TRUE
## 7 FALSE TRUE
## 8 FALSE TRUE
## 9 FALSE TRUE
## 10 FALSE TRUE
## # … with 90 more rows